Laboratory
What is the Antizyme ?
Polyamines are highly charged bioactive substances presented ubiquitously in species from bacteria to human. Polyamines are necessary for cell growth and are involved in highly diversified cellular functions such as cell division, apoptosis, autophagy, oxidative stress and ion channel activity. There are three major polyamines, putrescine, spermidine and spermine in the cells [1, 2]. Intracellular cellular polyamine concentration is highly regulated by the protein, “antizyme” [3-6] that is widely distributed from yeast to human [7]. Antizyme (AZ) is induced in response to increasing the concentration of intracellular polyamines through the polyamine-induced translational frameshifting mechanism [8]. AZ mRNA consists of two ORF, ORF1 and ORF2. In the low polyamine concentration, translation of ORF1 is terminated at stop codon “UGA” of ORF1 and short product is produced (Figure 1). But increasing cellular polyamine concentration, reading frame shifts +1 direction at the end of ORF1 (Figure 1 bottom column). In this case, following ORF1, ORF2 is translated and full length active product “antizyme” is produced [8, 9]. Induced AZ protein binds to ornithine decarboxylase (ODC) monomer, a key enzyme in polyamine biosynthesis and catalyze the conversion from ornithine to putrescine, and inhibit its activity. AZ-bound ODC is targeted to the 26S proteasome for degradation without ubiquitination [Figure 1, 10]. AZ also suppress polyamine uptake by inhibiting membrane polyamine transporter [Figure 1, 11, 12]. Thus AZ provides the negative feedback regulation of cellular polyamines. In addition, AZ is regulated by the protein, antizyme inhibitor (AZIN) that is homologous to ODC and can bind to AZ with higher affinity than ODC, but lacking the enzymatic activity [13, 14].
In mammals, cells express three members of AZ protein family, AZ1-3 (Table 1) [15]. AZ1 and AZ2 are distributed ubiquitously in most of tissues whereas AZ3 is testis specific [16-18]. Both AZ1 and AZ2 bind to ODC and accelerate its degradation in the cells [9, 19], but AZ3 has no activity for acceleration of ODC degradation [20]. The rate of ODC degradation by AZ1 is faster than that by AZ2 [19, 21]. Polyamine (putrescine) concentration of AZ1 knockdown cells is markedly increased, compared with that of AZ2 knockdown and control cells[26]. Therefore it is thought that AZ1 mainly regulates cellular polyamine concentration. On the other hand, although AZ2 is highly homologous to AZ1[21], it is considered that AZ2 is not backup of AZ1 because of some differences between each other. AZ2 was found as one of the genes upregulated in neuronal cells by the drug that induces seizure [23]. Nucleic acid sequence of AZ2 is evolutionally conserved higher than that of AZ1 [7]. AZ2 is localized mainly in the nucleus[26], and is phosphorylated in the cells [28]. Therefore we consider that AZ2 has specific function in the nuclear. We will mention about AZ2 specific function with its interacting protein we found very recently in this chapter.
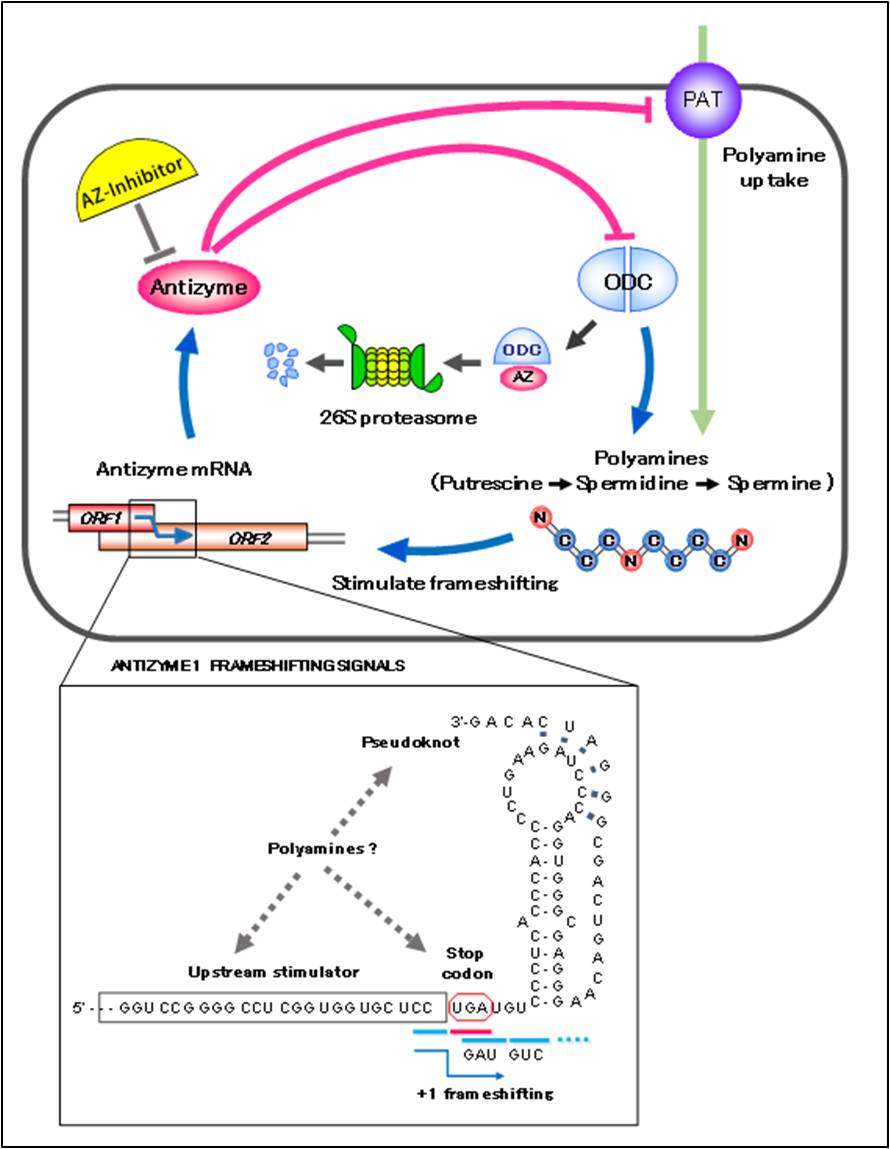
Figure 1 Negative feedback regulation of cellular polyamine by antizyme
Three cis-acting elements, UGA stop codon, upstream stimulater and pseudoknot structure, are known to be important for +1 frameshifting (bottom column) . Putrescine, spermidine and spermine are major polyamines in mammalian cell. Putrescine synthesized from ornithine by ODC could be metabolized to spermidine and spermine in the cells. PAT is a polyamine transporter which uptakes polyamines from outside of the cells.
Table 1 Characteristics of antizyme family
Newman, et al. reported that AZ1 has ability to accelerate the degradation of cyclin D1, one of the cell cycle regulatory protein family [28]. Cyclin D1 interact with cyclin dependent kinase (CDK), and accumulation of cyclin D1-CDK complex is important for cell cycle progression [29]. This protein is already known to be degraded by ubiquitin proteasome pathway [30]. They demonstrated that Az1 induction by polyamine or overexpression of AZ1 accelerates cyclin D1 degradation, and knockdown of AZ1 suppress it. Furthermore, in vitro experiment using purified cyclin D1, AZ1 and rabbit reticulocyte extracts as a source of 26S proteasome, AZ1 accelerated cyclin D1 degradation in a ATP-dependent manner. AZ1 could also degrade ubiquitin-deficient mutant of cyclin D1 in the cells [28]. In vitro size distribution analysis for binding between AZ1, cyclin D1, and ODC suggested that binding sites of AZ1 for cyclin D1 and ODC are not overlapped each other, and cyclin D1 binds to the N-terminus of AZ1 and ODC binds at the C-terminus, respectively. Binding affinity of AZ1 to cyclin D1 is 4-fold lower than that to ODC [31]. Although physiological significance is not clear, they showed that those three proteins form cyclin D1-AZ1-ODC ternary complex.
The oncogene Aurora A encodes a protein kinase that exerts essential roles in mitotic events and is important for induction of centrosome amplification [32]. Overexpression of Aurora A in many cancers induces aneuploidy, centrosome anomaly, poor prognosis and invasiveness [33, 34]. Aurora A is ubiquitinated by the E3 ubiquitin (Ub) ligase, anaphase-promoting complex/cyclosome (APC/C) that activated by both Cell-division cycle protein 20(Cdc20) and Cdh1, substrate-recognition subunit of APC/C, and is degraded by the proteasome [35, 36]. However Lim and Gopalan demonstrated that AZ1 could accelerates Aurora A degradation ubiquitin-independent manner, where Aurora-A kinase interacting protein 1 (AURKAIP1), a negative regulator of Aurora-A, enhances the binding of Az1 to Aurora-A, and facilitates the recognition of Aurora-A by the proteasome[37].
Mps1 is protein kinase required for centrosome duplication in regulating the spindle assembly checkpoint [38, 39]. Accumulation of Mps1 at centrosome causes aberrant centriole assembly [40, 41]. In fact in various tumor cells, centrosomal Mps1 pool is increased and that cause abnormal centrosome duplication [40]. Thus degradation of Mps1 is important for proper pool of Mps1 at centrosome. Although degradation of Mps1 is known to be mediated by the proteasome, amino acids residue 420-507 of the human Mps1 that is sufficient for its degradation does not contain APC/C recognition motifs, suggesting the commitment of Mps1 to ubiquitin-independent proteasomal degradation [41]. Kasbek et al. reported that AZ1 localizes to centrosomes and binds to Mps1 to controlling the levels of centrosomal Mps1 by accelerating the degradation of Mps1 [42]. Using fluorescent microscopy, they showed that centrosomal Mps1 level is depend on AZ1 expression and overexpression of AZ1 decrease the centrosome Mps1 level, conversely AZ1 knockdown by siRNA increase that. Furthermore, deletion of degradation signal of Mps1 abolished the regulation of centrosomal Mps1 level by AZ1. In addition, overexpressing AZIN in the cells to trap AZ1 and inhibit its function increased centrosomal Mps1 level. Thus the balance of AZ1 and Mps1 level in the centrosome is important for the centrosome duplication process.
P73 is a homologue of p53 and exist as two major forms, TAp73 or Delta-N (DN) p73. TAp73 is full-length form and exerts proapoptotic function, whereas DNp73, which is amino-terminal transactivation domain lacking form of p73 exhibits dominant-negative inhibitor activity for both p73 and p53, resulting in antiapoptotic properties[43]. Therefore, in the stress condition like DNA damage, reduction of DNp73 level is needed to execute apoptosis [44-46]. It has known that degradation of both TAp73 and DNp73 are mediated E3-ubiquitin ligase Itch in a proteasome dependent manner in normal condition [47]. However, in Itch decreased condition such as DNA damage by UV irradiation, stabilization of TAp73 was observed, but DNp73 was not [47]. Therefore it was considered that the degradation of TAp73 and DNp73 are regulated by different mechanisms. Dulloo et al. reported that reduction of DNp73 in the stress condition is due to the degradation of DNp73 by AZ1 mediated and ubiquitin-independent proteasomal pathway [48]. They showed that degradation of DNp73 could be induced genotoxic stresses such as UV irradiation and doxorubicin treatment. Inhibition of ubiquitin activating enzyme E1 by the inhibitor PYR41 could not block DNp73 degradation, indicated that this pathway occurs ubiquitin-independently. They demonstrated that polyamine induced AZ1 bind to DNp73 and accelerates its degradation. Interestingly, AZ1 mediated DNp73 degradation is dependent on transcription factor c-Jun which is activated by stress signals. Using overexpression and knocking down of AZ1, they also showed that even in the presence of c-Jun, AZ1 is necessary for genotoxic stress induced DNp73 degradation. Although it is not clear how c-Jun operates AZ1 expression, c-Jun may act upstream of polyamine biosynthesis pathway.
Thus, several proteins degraded by AZ1 mediated proteasome pathway are found, but AZ2 interacting protein or AZ2 mediated proteasomal degradation other than ODC has not been reported. We recently found two AZ2 interacting proteins and one of two was the protein accelerated its proteasomal degradation by AZ2 without ubiquitination (see next section).
It has known that degradation of c-Myc is mediated by ubiquitin-proteasome pathway, where c-Myc is phosphorylated at Thr-58 (pT58) and Ser-62 (pS62) by extracellular signal-regulated kinase, ERK and glycogen synthase kinase 3β, GSK-3β, respectably [56, 57]. After dephosphorylated at Ser-62 by protein phosphatase 2A, PP2A, pT58-c-Myc is ubiquitinated by E3-ubiquitin ligase Fbxw7 for proteasomal degradation [56, 58]. At first, AZ2 interacting protein identified by the comprehensive analysis mentioned above was not c-Myc but a protein that have basic region/helix-loop-helix/leucine zipper domain, and interacts with c-Myc (Murai et al., manuscript in preparation). However, in the process of analyzing interaction with AZ2, we found that AZ2 interacts with c-Myc in the cells by immuneprecipitation assay. Subcellular localization analysis of both proteins using fluorescent protein tags or antibodies conjugated fluorescent probe revealed that AZ2 co-localized with c-Myc in the nucleus. Treatment of proteasome inhibitor MG132 changes the nuclear co-localization of both proteins to nucleolar co-localization[22]. Overexpression of AZ2 or addition of polyamine in the cells accelerated c-Myc degradation, and knockdown of AZ2 with siRNA suppressed it. Furthermore, E1 inhibitor PYR-41 could not suppress AZ2 mediated proteasomal c-Myc degradation[22]. These results suggest that AZ2 mediated ubiquitin-independent nucleolar c-Myc degradation pathway other than ubiquitin-dependent one exist in the cells (Figure 2).
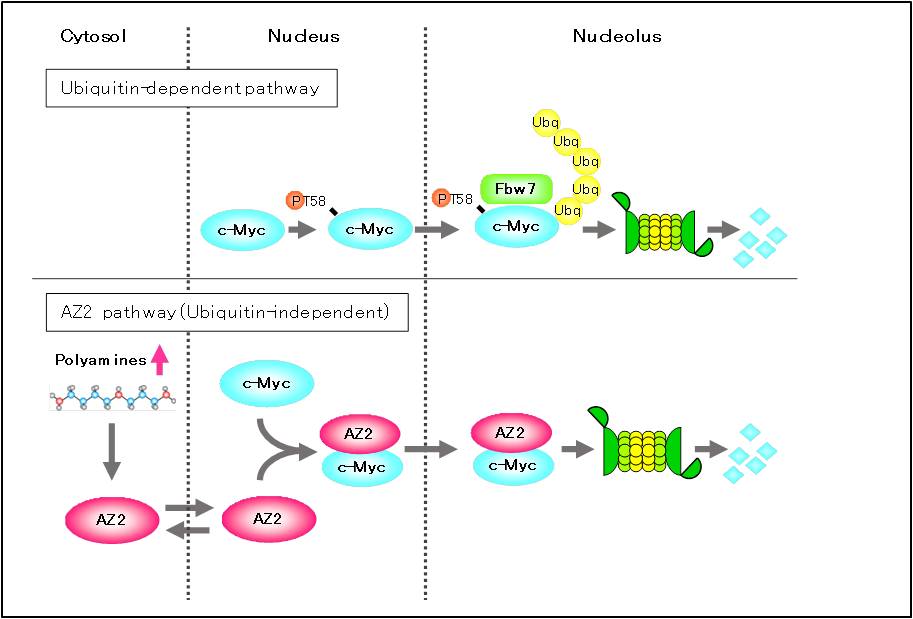
Figure 2 AZ2 mediated c-MYC degradation in the nucleolus
Tow distinct c-Myc degradation pathway exist in the cells. Increased polyamines induce expression of AZ2, and then AZ2 interacts with c-Myc in the nucleus and nucleolus to accelerate c-Myc degradation. It is thought that AZ2 pathway function under the stress condition (polyamine increased condition) such as glucose-free and hypoxia.
The proteins mentioned above are all known as the proteins degraded by ubiquitin-dependent proteasomal pathway other than ODC. It is not clear how antizyme mediated ubiquitin-independent degradation of these proteins are physiologically significant. Normally subcellular localization of ODC is mainly cytoplasm and at least not in the nucleolus even in the presence of MG132. In addition ODC is necessary for cell growth and the affinity of interaction between antizyme and ODC is high [59], in such condition, ODC probably occupies almost all antizyme in cytosol and antizyme hardly function for other antizyme interacting proteins [60]. In this context, because subcellular localization of both AZ2 and its interacting protein c-Myc are nucleus or nucleolus, cytosolic protein ODC could not interact with AZ2 in there. ODC is one of the c-Myc targeting proteins, AZ2 may function upstream of c-Myc especially under the stress condition such as glucose free and hypoxic condition [22]. Further studies are needed to elucidate the significance of antizyme-proteasome degradation pathway.
[2] Casero RA, Jr., Murray Stewart T, Pegg AE. Polyamine metabolism and cancer: treatments, challenges and opportunities. Nat Rev Cancer. 2018;18(11):681-95.
[3] Hayashi S, Murakami Y, Matsufuji S. Ornithine decarboxylase antizyme: a novel type of regulatory protein. Trends in biochemical sciences. 1996;21(1):27-30.
[4] Li X, Coffino P. Regulated degradation of ornithine decarboxylase requires interaction with the polyamine-inducible protein antizyme. Molecular and cellular biology. 1992;12(8):3556-62.
[5] Coffino P. Regulation of cellular polyamines by antizyme. Nature reviews Molecular cell biology. 2001;2(3):188-94.
[6] Heller JS, Fong WF, Canellakis ES. Induction of a protein inhibitor to ornithine decarboxylase by the end products of its reaction. Proc Natl Acad Sci U S A. 1976;73(6):1858-62.
[7] Ivanov IP, Gesteland RF, Atkins JF. Antizyme expression: a subversion of triplet decoding, which is remarkably conserved by evolution, is a sensor for an autoregulatory circuit. Nucleic Acids Res. 2000;28(17):3185-96.
[8] Matsufuji S, Matsufuji T, Miyazaki Y, Murakami Y, Atkins JF, Gesteland RF, et al. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell. 1995;80(1):51-60.
[9] Atkins JF, Loughran G, Bhatt PR, Firth AE, Baranov PV. Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use. Nucleic Acids Res. 2016;44(15):7007-78.
[10] Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, et al. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992;360(6404):597-9.
[11] Mitchell JL, Judd GG, Bareyal-Leyser A, Ling SY. Feedback repression of polyamine transport is mediated by antizyme in mammalian tissue-culture cells. The Biochemical journal. 1994;299 ( Pt 1):19-22.
[12] Suzuki T, He Y, Kashiwagi K, Murakami Y, Hayashi S, Igarashi K. Antizyme protects against abnormal accumulation and toxicity of polyamines in ornithine decarboxylase-overproducing cells. Proc Natl Acad Sci U S A. 1994;91(19):8930-4.
[13] Fujita K, Murakami Y, Hayashi S. A macromolecular inhibitor of the antizyme to ornithine decarboxylase. The Biochemical journal. 1982;204(3):647-52.
[14] Kitani T, Fujisawa H. Purification and characterization of antizyme inhibitor of ornithine decarboxylase from rat liver. Biochimica et biophysica acta. 1989;991(1):44-9.
[15] Ivanov IP, Gesteland RF, Atkins JF. SURVEY AND SUMMARY: Antizyme expression: a subversion of triplet decoding, which is remarkably conserved by evolution, is a sensor for an autoregulatory circuit. Nucleic Acids Research. 2000;28(17):3185-96.
[16] Ivanov IP, Gesteland RF, Atkins JF. A second mammalian antizyme: conservation of programmed ribosomal frameshifting. Genomics. 1998;52(2):119-29.
[17] Ivanov IP, Rohrwasser A, Terreros DA, Gesteland RF, Atkins JF. Discovery of a spermatogenesis stage-specific ornithine decarboxylase antizyme: antizyme 3. Proc Natl Acad Sci U S A. 2000;97(9):4808-13.
[18] Tosaka Y, Tanaka H, Yano Y, Masai K, Nozaki M, Yomogida K, et al. Identification and characterization of testis specific ornithine decarboxylase antizyme (OAZ-t) gene: expression in haploid germ cells and polyamine-induced frameshifting. Genes to cells : devoted to molecular & cellular mechanisms. 2000;5(4):265-76.
[19]Chen H, MacDonald A, Coffino P. Structural elements of antizymes 1 and 2 are required for proteasomal degradation of ornithine decarboxylase. J Biol Chem. 2002;277(48):45957-61.
[20] Snapir Z, Keren-Paz A, Bercovich Z, Kahana C. Antizyme 3 inhibits polyamine uptake and ornithine decarboxylase (ODC) activity, but does not stimulate ODC degradation. The Biochemical journal. 2009;419(1):99-103, 1 p following
[21] Zhu C, Lang DW, Coffino P. Antizyme2 is a negative regulator of ornithine decarboxylase and polyamine transport. J Biol Chem. 1999;274(37):26425-30.
[22] Murai N, Murakami Y, Tajima A, Matsufuji S. Novel ubiquitin-independent nucleolar c-Myc degradation pathway mediated by antizyme 2. Scientific reports. 2018;8(1):3005-.
[23] Kajiwara K, Nagawawa H, Shimizu-Nishikawa S, Ookuri T, Kimura M, Sugaya E. Molecular characterization of seizure-related genes isolated by differential screening. Biochemical and biophysical research communications. 1996;219(3):795-9.
[24] Murai N, Shimizu A, Murakami Y, Matsufuji S. Subcellular localization and phosphorylation of antizyme 2. Journal of cellular biochemistry. 2009;108(4):1012-21.
[25] Kawabata M, Imamura T, Miyazono K. Signal transduction by bone morphogenetic proteins. Cytokine & growth factor reviews. 1998;9(1):49-61.
[26] Bragdon B, Moseychuk O, Saldanha S, King D, Julian J, Nohe A. Bone morphogenetic proteins: a critical review. Cell Signal. 2011;23(4):609-20.
[27] Lin Y, Martin J, Gruendler C, Farley J, Meng X, Li BY, et al. A novel link between the proteasome pathway and the signal transduction pathway of the bone morphogenetic proteins (BMPs). BMC cell biology. 2002;3:15.
[28] Newman RM, Mobascher A, Mangold U, Koike C, Diah S, Schmidt M, et al. Antizyme targets cyclin D1 for degradation. A novel mechanism for cell growth repression. J Biol Chem. 2004;279(40):41504-11.
[29] Diehl JA. Cycling to cancer with cyclin D1. Cancer biology & therapy. 2002;1(3):226-31.
[30] Diehl JA, Zindy F, Sherr CJ. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes & development. 1997;11(8):957-72.
[31] Liu YC, Lee CY, Lin CL, Chen HY, Liu GY, Hung HC. Multifaceted interactions and regulation between antizyme and its interacting proteins cyclin D1, ornithine decarboxylase and antizyme inhibitor. Oncotarget. 2015.
[32] Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. The EMBO journal. 1998;17(11):3052-65.
[33] Sakakura C, Hagiwara A, Yasuoka R, Fujita Y, Nakanishi M, Masuda K, et al. Tumour-amplified kinase BTAK is amplified and overexpressed in gastric cancers with possible involvement in aneuploid formation. British journal of cancer. 2001;84(6):824-31.
[34] Buschhorn HM, Klein RR, Chambers SM, Hardy MC, Green S, Bearss D, et al. Aurora-A over-expression in high-grade PIN lesions and prostate cancer. The Prostate. 2005;64(4):341-6.
[35] Littlepage LE, Ruderman JV. Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes & development. 2002;16(17):2274-85.
[36] Taguchi S, Honda K, Sugiura K, Yamaguchi A, Furukawa K, Urano T. Degradation of human Aurora-A protein kinase is mediated by hCdh1. FEBS letters. 2002;519(1-3):59-65.
[37] Lim SK, Gopalan G. Antizyme1 mediates AURKAIP1-dependent degradation of Aurora-A. Oncogene. 2007;26(46):6593-603.
[38] Abrieu A, Magnaghi-Jaulin L, Kahana JA, Peter M, Castro A, Vigneron S, et al. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 2001;106(1):83-93.
[39] Stucke VM, Silljé HH, Arnaud L, Nigg EA. Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. The EMBO journal. 2002;21(7):1723-32.
[40] Kasbek C, Yang CH, Fisk HA. Mps1 as a link between centrosomes and genomic instability. Environmental and molecular mutagenesis. 2009;50(8):654-65.
[41] Kasbek C, Yang CH, Yusof AM, Chapman HM, Winey M, Fisk HA. Preventing the degradation of mps1 at centrosomes is sufficient to cause centrosome reduplication in human cells. Mol Biol Cell. 2007;18(11):4457-69.
[42] Kasbek C, Yang CH, Fisk HA. Antizyme restrains centrosome amplification by regulating the accumulation of Mps1 at centrosomes. Mol Biol Cell. 2010;21(22):3878-89.
[43] Melino G, De Laurenzi V, Vousden KH. p73: Friend or foe in tumorigenesis. Nat Rev Cancer. 2002;2(8):605-15.
[44] Irwin MS, Kondo K, Marin MC, Cheng LS, Hahn WC, Kaelin WG, Jr. Chemosensitivity linked to p73 function. Cancer cell. 2003;3(4):403-10.
[45] Lin KW, Nam SY, Toh WH, Dulloo I, Sabapathy K. Multiple stress signals induce p73beta accumulation. Neoplasia (New York, NY). 2004;6(5):546-57.
[46] Maisse C, Munarriz E, Barcaroli D, Melino G, De Laurenzi V. DNA damage induces the rapid and selective degradation of the DeltaNp73 isoform, allowing apoptosis to occur. Cell death and differentiation. 2004;11(6):685-7.
[47] Rossi M, De Laurenzi V, Munarriz E, Green DR, Liu YC, Vousden KH, et al. The ubiquitin-protein ligase Itch regulates p73 stability. The EMBO journal. 2005;24(4):836-48.
[48] Dulloo I, Gopalan G, Melino G, Sabapathy K. The antiapoptotic DeltaNp73 is degraded in a c-Jun-dependent manner upon genotoxic stress through the antizyme-mediated pathway. Proc Natl Acad Sci U S A. 2010;107(11):4902-7.
[49] Ramos-Molina B, Lopez-Contreras AJ, Cremades A, Penafiel R. Differential expression of ornithine decarboxylase antizyme inhibitors and antizymes in rodent tissues and human cell lines. Amino Acids. 2012;42(2-3):539-47.
[50] Watson JA, Fang M, Lowenstein JM. Tricarballylate and hydroxycitrate: substrate and inhibitor of ATP: citrate oxaloacetate lyase. Archives of biochemistry and biophysics. 1969;135(1):209-17.
[51] Chypre M, Zaidi N, Smans K. ATP-citrate lyase: a mini-review. Biochemical and biophysical research communications. 2012;422(1):1-4.
[52] Tajima A, Murai N, Murakami Y, Iwamoto T, Migita T, Matsufuji S. Polyamine regulating protein antizyme binds to ATP citrate lyase to accelerate acetyl-CoA production in cancer cells. Biochemical and biophysical research communications. 2016.
[53] Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8(12):976-90.
[54] van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10(4):301-9.
[55] Bello-Fernandez C, Packham G, Cleveland JL. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci U S A. 1993;90(16):7804-8.
[56] Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. The EMBO journal. 2004;23(10):2116-25.
[57] Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci U S A. 2004;101(24):9085-90.
[58] Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6(4):308-18.
[59] Liu YC, Hsu DH, Huang CL, Liu YL, Liu GY, Hung HC. Determinants of the differential antizyme-binding affinity of ornithine decarboxylase. PLoS One. 2011;6(11):e26835.
[60] Bercovich Z, Snapir Z, Keren-Paz A, Kahana C. Antizyme affects cell proliferation and viability solely through regulating cellular polyamines. J Biol Chem. 2011;286(39):33778-83.
In mammals, cells express three members of AZ protein family, AZ1-3 (Table 1) [15]. AZ1 and AZ2 are distributed ubiquitously in most of tissues whereas AZ3 is testis specific [16-18]. Both AZ1 and AZ2 bind to ODC and accelerate its degradation in the cells [9, 19], but AZ3 has no activity for acceleration of ODC degradation [20]. The rate of ODC degradation by AZ1 is faster than that by AZ2 [19, 21]. Polyamine (putrescine) concentration of AZ1 knockdown cells is markedly increased, compared with that of AZ2 knockdown and control cells[26]. Therefore it is thought that AZ1 mainly regulates cellular polyamine concentration. On the other hand, although AZ2 is highly homologous to AZ1[21], it is considered that AZ2 is not backup of AZ1 because of some differences between each other. AZ2 was found as one of the genes upregulated in neuronal cells by the drug that induces seizure [23]. Nucleic acid sequence of AZ2 is evolutionally conserved higher than that of AZ1 [7]. AZ2 is localized mainly in the nucleus[26], and is phosphorylated in the cells [28]. Therefore we consider that AZ2 has specific function in the nuclear. We will mention about AZ2 specific function with its interacting protein we found very recently in this chapter.

Figure 1 Negative feedback regulation of cellular polyamine by antizyme
Three cis-acting elements, UGA stop codon, upstream stimulater and pseudoknot structure, are known to be important for +1 frameshifting (bottom column) . Putrescine, spermidine and spermine are major polyamines in mammalian cell. Putrescine synthesized from ornithine by ODC could be metabolized to spermidine and spermine in the cells. PAT is a polyamine transporter which uptakes polyamines from outside of the cells.
Table 1 Characteristics of antizyme family
| AZ1 | AZ2 | AZ3 | |
| Species distribution | Yeast-mammals | Vertebrates | Mammals |
| Evolutional conservation | Lower | Higher | Lower |
| Tissue distribution | Whole body | Whole body | Testis |
| mRNA Expression | High | Low | Low |
| Cellular distribution | Cytoplasm and Nucleus |
Mainly | Spermatid and sperm |
| Induction by +1 frameshifting |
+ | + | + |
| ODC binding and inhibition | + | + | + |
| Acceleration of ODC degradation | |||
| in vivo | + | + | − |
| in vitro | + | − | − |
| Inhibition of polyamine uptake | + | + | + |
| AZ inhibitor binding | + | + | + |
| Phenotype of knockout mice | unreported | unreported | Male infertility |
Antizyme interacting proteins and ubiquitin-independent proteasomasl degradation.
1. Antizyme 1 interacting proteins
It had been considered that ODC is the only protein degraded through AZ mediated ubiquitin-dependent proteasomal degradation system. However, recently several AZ1 interacting proteins other than ODC have reported (Table 2) Although it has already been reported that those proteins are degraded by the ubiquitin-proteasome pathway, AZ1 could also accelerates those degradation without ubiquitination (Table 1, 2, Figure 1). Smad1, which is involved in bone morphogenetic proteins (BMP) signaling pathway [25, 26], is the first reported protein that interacts AZ1 other than ODC [27]. In this case, newly-synthesized HsN3, which is β-subunit for 20S proteasome, forms ternary complex with AZ1 and smad1. This complex may binds to 20S proteasome, and next 15S complex is docked on 20S, and then smad1 is degraded by the 26S proteasome.Table 2 The proteins degraded by antizyme mediated ubiquitin-independent proteasomal pathway
| Protein names | Protein function | Biological process | Characteristics of degradation |
| ODC | Synthesis of putrescine from ornithine | Polyamine metabolism | AZ1 or AZ2 binds to ODC monomer that is targeted to the 26S proteasome [10, 19] |
| Smad1 | Binding to smad4 Regulation of transcription | BMP signaling pathway | Formation of HsN3-Smad1-AZ1 ternary complex [27]. |
| CyclinD1 | Binding to CDK | Cell cycle | AZ1 binds to Cyclin D1 and accelerates its degradation In vitro and in vivo [28]. |
| Aurora A | Protein Kinase | Mitotic events | Formation of AUKAIP-1-AZ1-Aurora-A ternary complex, and that is target to the proteasome in vivo [41]. |
| Msp1 | Protein Kinase | Cell divition | AZ1 affects the level of Msp1at the centrosome [46]. |
| DNp73 | Inhibition of both p73 and p53 | Apoptosis | c-Jun dependent DNp73 degradation mediated by AZ1 in vivo [48]. |
| c-Myc | Transcription factor | Cell growth Differentiation survival Apoptosis | AZ2 binds to c-Myc and accelerates its degradation in vivo [22]. |
Newman, et al. reported that AZ1 has ability to accelerate the degradation of cyclin D1, one of the cell cycle regulatory protein family [28]. Cyclin D1 interact with cyclin dependent kinase (CDK), and accumulation of cyclin D1-CDK complex is important for cell cycle progression [29]. This protein is already known to be degraded by ubiquitin proteasome pathway [30]. They demonstrated that Az1 induction by polyamine or overexpression of AZ1 accelerates cyclin D1 degradation, and knockdown of AZ1 suppress it. Furthermore, in vitro experiment using purified cyclin D1, AZ1 and rabbit reticulocyte extracts as a source of 26S proteasome, AZ1 accelerated cyclin D1 degradation in a ATP-dependent manner. AZ1 could also degrade ubiquitin-deficient mutant of cyclin D1 in the cells [28]. In vitro size distribution analysis for binding between AZ1, cyclin D1, and ODC suggested that binding sites of AZ1 for cyclin D1 and ODC are not overlapped each other, and cyclin D1 binds to the N-terminus of AZ1 and ODC binds at the C-terminus, respectively. Binding affinity of AZ1 to cyclin D1 is 4-fold lower than that to ODC [31]. Although physiological significance is not clear, they showed that those three proteins form cyclin D1-AZ1-ODC ternary complex.
The oncogene Aurora A encodes a protein kinase that exerts essential roles in mitotic events and is important for induction of centrosome amplification [32]. Overexpression of Aurora A in many cancers induces aneuploidy, centrosome anomaly, poor prognosis and invasiveness [33, 34]. Aurora A is ubiquitinated by the E3 ubiquitin (Ub) ligase, anaphase-promoting complex/cyclosome (APC/C) that activated by both Cell-division cycle protein 20(Cdc20) and Cdh1, substrate-recognition subunit of APC/C, and is degraded by the proteasome [35, 36]. However Lim and Gopalan demonstrated that AZ1 could accelerates Aurora A degradation ubiquitin-independent manner, where Aurora-A kinase interacting protein 1 (AURKAIP1), a negative regulator of Aurora-A, enhances the binding of Az1 to Aurora-A, and facilitates the recognition of Aurora-A by the proteasome[37].
Mps1 is protein kinase required for centrosome duplication in regulating the spindle assembly checkpoint [38, 39]. Accumulation of Mps1 at centrosome causes aberrant centriole assembly [40, 41]. In fact in various tumor cells, centrosomal Mps1 pool is increased and that cause abnormal centrosome duplication [40]. Thus degradation of Mps1 is important for proper pool of Mps1 at centrosome. Although degradation of Mps1 is known to be mediated by the proteasome, amino acids residue 420-507 of the human Mps1 that is sufficient for its degradation does not contain APC/C recognition motifs, suggesting the commitment of Mps1 to ubiquitin-independent proteasomal degradation [41]. Kasbek et al. reported that AZ1 localizes to centrosomes and binds to Mps1 to controlling the levels of centrosomal Mps1 by accelerating the degradation of Mps1 [42]. Using fluorescent microscopy, they showed that centrosomal Mps1 level is depend on AZ1 expression and overexpression of AZ1 decrease the centrosome Mps1 level, conversely AZ1 knockdown by siRNA increase that. Furthermore, deletion of degradation signal of Mps1 abolished the regulation of centrosomal Mps1 level by AZ1. In addition, overexpressing AZIN in the cells to trap AZ1 and inhibit its function increased centrosomal Mps1 level. Thus the balance of AZ1 and Mps1 level in the centrosome is important for the centrosome duplication process.
P73 is a homologue of p53 and exist as two major forms, TAp73 or Delta-N (DN) p73. TAp73 is full-length form and exerts proapoptotic function, whereas DNp73, which is amino-terminal transactivation domain lacking form of p73 exhibits dominant-negative inhibitor activity for both p73 and p53, resulting in antiapoptotic properties[43]. Therefore, in the stress condition like DNA damage, reduction of DNp73 level is needed to execute apoptosis [44-46]. It has known that degradation of both TAp73 and DNp73 are mediated E3-ubiquitin ligase Itch in a proteasome dependent manner in normal condition [47]. However, in Itch decreased condition such as DNA damage by UV irradiation, stabilization of TAp73 was observed, but DNp73 was not [47]. Therefore it was considered that the degradation of TAp73 and DNp73 are regulated by different mechanisms. Dulloo et al. reported that reduction of DNp73 in the stress condition is due to the degradation of DNp73 by AZ1 mediated and ubiquitin-independent proteasomal pathway [48]. They showed that degradation of DNp73 could be induced genotoxic stresses such as UV irradiation and doxorubicin treatment. Inhibition of ubiquitin activating enzyme E1 by the inhibitor PYR41 could not block DNp73 degradation, indicated that this pathway occurs ubiquitin-independently. They demonstrated that polyamine induced AZ1 bind to DNp73 and accelerates its degradation. Interestingly, AZ1 mediated DNp73 degradation is dependent on transcription factor c-Jun which is activated by stress signals. Using overexpression and knocking down of AZ1, they also showed that even in the presence of c-Jun, AZ1 is necessary for genotoxic stress induced DNp73 degradation. Although it is not clear how c-Jun operates AZ1 expression, c-Jun may act upstream of polyamine biosynthesis pathway.
Thus, several proteins degraded by AZ1 mediated proteasome pathway are found, but AZ2 interacting protein or AZ2 mediated proteasomal degradation other than ODC has not been reported. We recently found two AZ2 interacting proteins and one of two was the protein accelerated its proteasomal degradation by AZ2 without ubiquitination (see next section).
2. Antizyme 2 interacting proteins
As mentioned above, AZ2 also binds to ODC and accelerates its degradation in the cells [5]. However, we have considered that AZ2 has specific function other than AZ1 because of the differences such as nuclear localization [22, 24], highly gene conservation between species [16] and high expression in neuronal cells [49]. We performed comprehensive analysis of AZ2 interacting protein using two hybrid technique. Two AZ2 interacting proteins were identified. One is ATP citrate lyase (ACLY) which is the enzyme catalyzing acetyl-CoA production in cytosol [50], and related to lipid anabolism and acetylation of cellular components [51]. We found that ACLY binds not only to AZ2 but also to AZ1 by immunoprecipitation assay [52]. Degradation assay for ACLY was performed in expectation of ubiquitin-independent proteasomal degradation, however AZs have no ability to accelerate ACLY degradation. Surprisingly, AZ1 and AZ2 activate catalytic activity of ACLY [52]. The other is proto-oncogene c-Myc that is transcription factor having a basic region/helix-loop-helix/leucine zipper domain, and form heterodimer with Max for DNA binding [53, 54]. c-Myc exerts function as master regulator of variety cellular processes such as cell growth, differentiation, survival and apoptosis [54]. In cell growth, c-Myc targets ODC gene [55] and promote synthesis of polyamine that is important for stabilization of nucleic acids, transcription, translation and +1 frameshifting on AZ mRNA [2].It has known that degradation of c-Myc is mediated by ubiquitin-proteasome pathway, where c-Myc is phosphorylated at Thr-58 (pT58) and Ser-62 (pS62) by extracellular signal-regulated kinase, ERK and glycogen synthase kinase 3β, GSK-3β, respectably [56, 57]. After dephosphorylated at Ser-62 by protein phosphatase 2A, PP2A, pT58-c-Myc is ubiquitinated by E3-ubiquitin ligase Fbxw7 for proteasomal degradation [56, 58]. At first, AZ2 interacting protein identified by the comprehensive analysis mentioned above was not c-Myc but a protein that have basic region/helix-loop-helix/leucine zipper domain, and interacts with c-Myc (Murai et al., manuscript in preparation). However, in the process of analyzing interaction with AZ2, we found that AZ2 interacts with c-Myc in the cells by immuneprecipitation assay. Subcellular localization analysis of both proteins using fluorescent protein tags or antibodies conjugated fluorescent probe revealed that AZ2 co-localized with c-Myc in the nucleus. Treatment of proteasome inhibitor MG132 changes the nuclear co-localization of both proteins to nucleolar co-localization[22]. Overexpression of AZ2 or addition of polyamine in the cells accelerated c-Myc degradation, and knockdown of AZ2 with siRNA suppressed it. Furthermore, E1 inhibitor PYR-41 could not suppress AZ2 mediated proteasomal c-Myc degradation[22]. These results suggest that AZ2 mediated ubiquitin-independent nucleolar c-Myc degradation pathway other than ubiquitin-dependent one exist in the cells (Figure 2).

Figure 2 AZ2 mediated c-MYC degradation in the nucleolus
Tow distinct c-Myc degradation pathway exist in the cells. Increased polyamines induce expression of AZ2, and then AZ2 interacts with c-Myc in the nucleus and nucleolus to accelerate c-Myc degradation. It is thought that AZ2 pathway function under the stress condition (polyamine increased condition) such as glucose-free and hypoxia.
The proteins mentioned above are all known as the proteins degraded by ubiquitin-dependent proteasomal pathway other than ODC. It is not clear how antizyme mediated ubiquitin-independent degradation of these proteins are physiologically significant. Normally subcellular localization of ODC is mainly cytoplasm and at least not in the nucleolus even in the presence of MG132. In addition ODC is necessary for cell growth and the affinity of interaction between antizyme and ODC is high [59], in such condition, ODC probably occupies almost all antizyme in cytosol and antizyme hardly function for other antizyme interacting proteins [60]. In this context, because subcellular localization of both AZ2 and its interacting protein c-Myc are nucleus or nucleolus, cytosolic protein ODC could not interact with AZ2 in there. ODC is one of the c-Myc targeting proteins, AZ2 may function upstream of c-Myc especially under the stress condition such as glucose free and hypoxic condition [22]. Further studies are needed to elucidate the significance of antizyme-proteasome degradation pathway.
References
[1] Pegg AE, Casero RA, Jr. Current status of the polyamine research field. Methods in molecular biology (Clifton, NJ). 2011;720:3-35.[2] Casero RA, Jr., Murray Stewart T, Pegg AE. Polyamine metabolism and cancer: treatments, challenges and opportunities. Nat Rev Cancer. 2018;18(11):681-95.
[3] Hayashi S, Murakami Y, Matsufuji S. Ornithine decarboxylase antizyme: a novel type of regulatory protein. Trends in biochemical sciences. 1996;21(1):27-30.
[4] Li X, Coffino P. Regulated degradation of ornithine decarboxylase requires interaction with the polyamine-inducible protein antizyme. Molecular and cellular biology. 1992;12(8):3556-62.
[5] Coffino P. Regulation of cellular polyamines by antizyme. Nature reviews Molecular cell biology. 2001;2(3):188-94.
[6] Heller JS, Fong WF, Canellakis ES. Induction of a protein inhibitor to ornithine decarboxylase by the end products of its reaction. Proc Natl Acad Sci U S A. 1976;73(6):1858-62.
[7] Ivanov IP, Gesteland RF, Atkins JF. Antizyme expression: a subversion of triplet decoding, which is remarkably conserved by evolution, is a sensor for an autoregulatory circuit. Nucleic Acids Res. 2000;28(17):3185-96.
[8] Matsufuji S, Matsufuji T, Miyazaki Y, Murakami Y, Atkins JF, Gesteland RF, et al. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell. 1995;80(1):51-60.
[9] Atkins JF, Loughran G, Bhatt PR, Firth AE, Baranov PV. Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use. Nucleic Acids Res. 2016;44(15):7007-78.
[10] Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, et al. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992;360(6404):597-9.
[11] Mitchell JL, Judd GG, Bareyal-Leyser A, Ling SY. Feedback repression of polyamine transport is mediated by antizyme in mammalian tissue-culture cells. The Biochemical journal. 1994;299 ( Pt 1):19-22.
[12] Suzuki T, He Y, Kashiwagi K, Murakami Y, Hayashi S, Igarashi K. Antizyme protects against abnormal accumulation and toxicity of polyamines in ornithine decarboxylase-overproducing cells. Proc Natl Acad Sci U S A. 1994;91(19):8930-4.
[13] Fujita K, Murakami Y, Hayashi S. A macromolecular inhibitor of the antizyme to ornithine decarboxylase. The Biochemical journal. 1982;204(3):647-52.
[14] Kitani T, Fujisawa H. Purification and characterization of antizyme inhibitor of ornithine decarboxylase from rat liver. Biochimica et biophysica acta. 1989;991(1):44-9.
[15] Ivanov IP, Gesteland RF, Atkins JF. SURVEY AND SUMMARY: Antizyme expression: a subversion of triplet decoding, which is remarkably conserved by evolution, is a sensor for an autoregulatory circuit. Nucleic Acids Research. 2000;28(17):3185-96.
[16] Ivanov IP, Gesteland RF, Atkins JF. A second mammalian antizyme: conservation of programmed ribosomal frameshifting. Genomics. 1998;52(2):119-29.
[17] Ivanov IP, Rohrwasser A, Terreros DA, Gesteland RF, Atkins JF. Discovery of a spermatogenesis stage-specific ornithine decarboxylase antizyme: antizyme 3. Proc Natl Acad Sci U S A. 2000;97(9):4808-13.
[18] Tosaka Y, Tanaka H, Yano Y, Masai K, Nozaki M, Yomogida K, et al. Identification and characterization of testis specific ornithine decarboxylase antizyme (OAZ-t) gene: expression in haploid germ cells and polyamine-induced frameshifting. Genes to cells : devoted to molecular & cellular mechanisms. 2000;5(4):265-76.
[19]Chen H, MacDonald A, Coffino P. Structural elements of antizymes 1 and 2 are required for proteasomal degradation of ornithine decarboxylase. J Biol Chem. 2002;277(48):45957-61.
[20] Snapir Z, Keren-Paz A, Bercovich Z, Kahana C. Antizyme 3 inhibits polyamine uptake and ornithine decarboxylase (ODC) activity, but does not stimulate ODC degradation. The Biochemical journal. 2009;419(1):99-103, 1 p following
[21] Zhu C, Lang DW, Coffino P. Antizyme2 is a negative regulator of ornithine decarboxylase and polyamine transport. J Biol Chem. 1999;274(37):26425-30.
[22] Murai N, Murakami Y, Tajima A, Matsufuji S. Novel ubiquitin-independent nucleolar c-Myc degradation pathway mediated by antizyme 2. Scientific reports. 2018;8(1):3005-.
[23] Kajiwara K, Nagawawa H, Shimizu-Nishikawa S, Ookuri T, Kimura M, Sugaya E. Molecular characterization of seizure-related genes isolated by differential screening. Biochemical and biophysical research communications. 1996;219(3):795-9.
[24] Murai N, Shimizu A, Murakami Y, Matsufuji S. Subcellular localization and phosphorylation of antizyme 2. Journal of cellular biochemistry. 2009;108(4):1012-21.
[25] Kawabata M, Imamura T, Miyazono K. Signal transduction by bone morphogenetic proteins. Cytokine & growth factor reviews. 1998;9(1):49-61.
[26] Bragdon B, Moseychuk O, Saldanha S, King D, Julian J, Nohe A. Bone morphogenetic proteins: a critical review. Cell Signal. 2011;23(4):609-20.
[27] Lin Y, Martin J, Gruendler C, Farley J, Meng X, Li BY, et al. A novel link between the proteasome pathway and the signal transduction pathway of the bone morphogenetic proteins (BMPs). BMC cell biology. 2002;3:15.
[28] Newman RM, Mobascher A, Mangold U, Koike C, Diah S, Schmidt M, et al. Antizyme targets cyclin D1 for degradation. A novel mechanism for cell growth repression. J Biol Chem. 2004;279(40):41504-11.
[29] Diehl JA. Cycling to cancer with cyclin D1. Cancer biology & therapy. 2002;1(3):226-31.
[30] Diehl JA, Zindy F, Sherr CJ. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes & development. 1997;11(8):957-72.
[31] Liu YC, Lee CY, Lin CL, Chen HY, Liu GY, Hung HC. Multifaceted interactions and regulation between antizyme and its interacting proteins cyclin D1, ornithine decarboxylase and antizyme inhibitor. Oncotarget. 2015.
[32] Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. The EMBO journal. 1998;17(11):3052-65.
[33] Sakakura C, Hagiwara A, Yasuoka R, Fujita Y, Nakanishi M, Masuda K, et al. Tumour-amplified kinase BTAK is amplified and overexpressed in gastric cancers with possible involvement in aneuploid formation. British journal of cancer. 2001;84(6):824-31.
[34] Buschhorn HM, Klein RR, Chambers SM, Hardy MC, Green S, Bearss D, et al. Aurora-A over-expression in high-grade PIN lesions and prostate cancer. The Prostate. 2005;64(4):341-6.
[35] Littlepage LE, Ruderman JV. Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes & development. 2002;16(17):2274-85.
[36] Taguchi S, Honda K, Sugiura K, Yamaguchi A, Furukawa K, Urano T. Degradation of human Aurora-A protein kinase is mediated by hCdh1. FEBS letters. 2002;519(1-3):59-65.
[37] Lim SK, Gopalan G. Antizyme1 mediates AURKAIP1-dependent degradation of Aurora-A. Oncogene. 2007;26(46):6593-603.
[38] Abrieu A, Magnaghi-Jaulin L, Kahana JA, Peter M, Castro A, Vigneron S, et al. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 2001;106(1):83-93.
[39] Stucke VM, Silljé HH, Arnaud L, Nigg EA. Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. The EMBO journal. 2002;21(7):1723-32.
[40] Kasbek C, Yang CH, Fisk HA. Mps1 as a link between centrosomes and genomic instability. Environmental and molecular mutagenesis. 2009;50(8):654-65.
[41] Kasbek C, Yang CH, Yusof AM, Chapman HM, Winey M, Fisk HA. Preventing the degradation of mps1 at centrosomes is sufficient to cause centrosome reduplication in human cells. Mol Biol Cell. 2007;18(11):4457-69.
[42] Kasbek C, Yang CH, Fisk HA. Antizyme restrains centrosome amplification by regulating the accumulation of Mps1 at centrosomes. Mol Biol Cell. 2010;21(22):3878-89.
[43] Melino G, De Laurenzi V, Vousden KH. p73: Friend or foe in tumorigenesis. Nat Rev Cancer. 2002;2(8):605-15.
[44] Irwin MS, Kondo K, Marin MC, Cheng LS, Hahn WC, Kaelin WG, Jr. Chemosensitivity linked to p73 function. Cancer cell. 2003;3(4):403-10.
[45] Lin KW, Nam SY, Toh WH, Dulloo I, Sabapathy K. Multiple stress signals induce p73beta accumulation. Neoplasia (New York, NY). 2004;6(5):546-57.
[46] Maisse C, Munarriz E, Barcaroli D, Melino G, De Laurenzi V. DNA damage induces the rapid and selective degradation of the DeltaNp73 isoform, allowing apoptosis to occur. Cell death and differentiation. 2004;11(6):685-7.
[47] Rossi M, De Laurenzi V, Munarriz E, Green DR, Liu YC, Vousden KH, et al. The ubiquitin-protein ligase Itch regulates p73 stability. The EMBO journal. 2005;24(4):836-48.
[48] Dulloo I, Gopalan G, Melino G, Sabapathy K. The antiapoptotic DeltaNp73 is degraded in a c-Jun-dependent manner upon genotoxic stress through the antizyme-mediated pathway. Proc Natl Acad Sci U S A. 2010;107(11):4902-7.
[49] Ramos-Molina B, Lopez-Contreras AJ, Cremades A, Penafiel R. Differential expression of ornithine decarboxylase antizyme inhibitors and antizymes in rodent tissues and human cell lines. Amino Acids. 2012;42(2-3):539-47.
[50] Watson JA, Fang M, Lowenstein JM. Tricarballylate and hydroxycitrate: substrate and inhibitor of ATP: citrate oxaloacetate lyase. Archives of biochemistry and biophysics. 1969;135(1):209-17.
[51] Chypre M, Zaidi N, Smans K. ATP-citrate lyase: a mini-review. Biochemical and biophysical research communications. 2012;422(1):1-4.
[52] Tajima A, Murai N, Murakami Y, Iwamoto T, Migita T, Matsufuji S. Polyamine regulating protein antizyme binds to ATP citrate lyase to accelerate acetyl-CoA production in cancer cells. Biochemical and biophysical research communications. 2016.
[53] Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8(12):976-90.
[54] van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10(4):301-9.
[55] Bello-Fernandez C, Packham G, Cleveland JL. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci U S A. 1993;90(16):7804-8.
[56] Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. The EMBO journal. 2004;23(10):2116-25.
[57] Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci U S A. 2004;101(24):9085-90.
[58] Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol. 2004;6(4):308-18.
[59] Liu YC, Hsu DH, Huang CL, Liu YL, Liu GY, Hung HC. Determinants of the differential antizyme-binding affinity of ornithine decarboxylase. PLoS One. 2011;6(11):e26835.
[60] Bercovich Z, Snapir Z, Keren-Paz A, Kahana C. Antizyme affects cell proliferation and viability solely through regulating cellular polyamines. J Biol Chem. 2011;286(39):33778-83.